Adjunct Assistant Professor
Ph.D. Plant Biology University of California at Berkeley, 2012
B.A. Natural Sciences New College of Florida, 1998
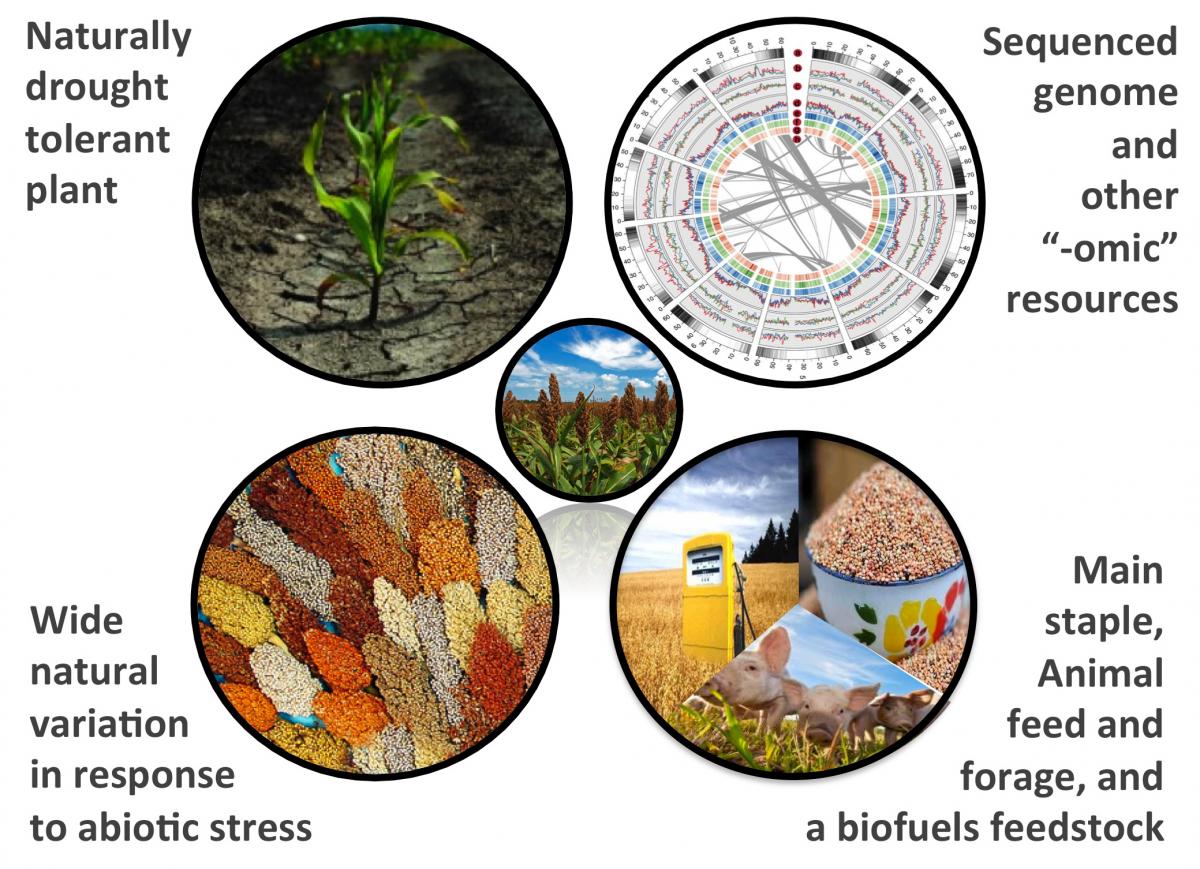
Our laboratory investigates the effects of drought and other abiotic stresses on the microbiomes associated with Sorghum bicolor and other grass species. With the world population expected to reach 9 billion by 2050, it is estimated that the global food supply will need to increase by 70 percent to meet rapidly rising demand. Changes in the global climate may well compound this challenge, as predicted increases in environmental stresses, such as drought and high-salinity, are expected to reduce crop productivity. Recent studies demonstrated that microbial symbionts of crop plants are capable of enhancing the abiotic stress tolerance of their host. However, only a tiny fraction of plant microbiomes have been uncovered and evaluated. Thus, research and new tools are needed to develop a better understanding of the interrelationship between crop plant abiotic stress tolerance and crop plant microbiomes.
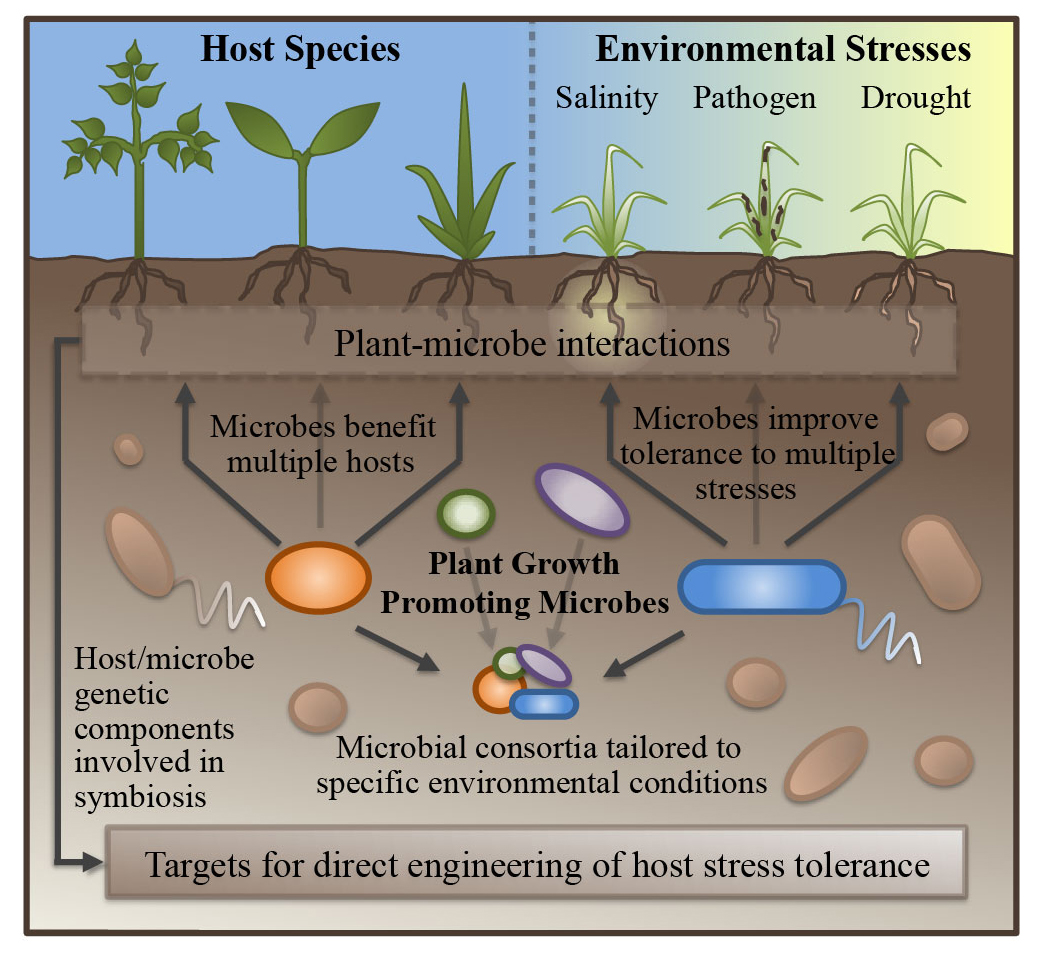
Our lab is also interested in understanding the role of plant growth promoting microbes (PGPM) in improving abiotic stress response in their plant hosts. Using a variety of culture and genomics-based approaches, we are working to uncover the host machinery manipulated by several PGPM species, which in turn may serve as potential targets for direct crop improvement through conventional breeding practices.-Additionally, the lab works on developing statistical methods and computational tools to investigate plant-associated microbiomes and stress response. These tools will be used to enable cross-species and cross-environment comparitive analyses, and a more holistic understanding of the plant and its microbial community.
Natural variaton in drought response in the phytobiomes of Sorghum bicolor and other crop species
Plant-associated microbial communities play an essential role in determining the phenotypic output of their hosts. Some of these bacteria and fungi confer benefit to crop species through increasing nutrient and resource uptake efficiency, out-competing plant pathogens, and improving abiotic stress response. Despite much research, only a tiny fraction of plant microbiomes have been uncovered and evaluated, and many of the rules governing microbial community recruitment to the host microbiome remain unknown, especially in atypically environments such as those governed by environmental stress. Drought stress is arguably the most critical environmental stress for crop production in many parts of the world and will increasingly become a greater factor with climate change; many microbial species have been shown to be capable of improving drought response in a variety of crop species. Sorghum bicolor, a crop with both feedstock and biofuels potential, is frequently grown in dry climates with frequent periods of drought.
Sorghum as a model for drought tolerance research
We are interested in addressing the following questions related to phytobiomes, drought stress and Sorghum bicolor: 1) What are the general and species-specific plant microbiome responses to abiotic stress across the grasses? Recent work in other plant systems has demonstrated that host genetics can play a role in selecting for specific microbial communities, but the extent to which the plant microbiome differs under abiotic stress between diverse grass species grown within the same soil has yet to be explored. 2) When and how do plant microbiomes respond to drought? The proposed work will uncover temporal changes in bacterial community structure in Sorghum bicolor as it acclimates to and recovers from drought, identifying key community members that increase and decrease in abundance during environmental stress. 3) How does species-species interaction within microbial communities affect relative root colonization efficiency under drought, and to what extent do root endophytes of sorghum effectively colonize other grass species? A novel high-throughput pooling strategy will be used to screen strains from a microbial library for their relative ability to colonize sorghum and to promote plant growth under drought conditions.
Plant growth promoting microbes and abiotic stress response in crop species
A large variety of plant growth promoting microbes (PGPM) have been shown to be capable of reducing abiotic stress response in their plant hosts, though the mechanisms through which they act are still being explored and established. In addition to a number of characterized strategies of growth promotion, including improved nutrient exchange, production of phytohormones, and competition with pathogens, recent research continues to unveil novel pathways through which PGPM benefit their hosts. Using genomics-based approaches, we are working to uncover the host machinery manipulated by several PGPM species in Sorghum and other species.
Mechanisms of plant growth promotion by microbial symbionts
Plant Microbiome Comutational Tool Development
A number of challenges exist for cross-study comparative analysis of microbiome datasets. Several well-documented pipelines exist for the analysis of individual datasets, such as QIIME and Mothur (Schloss et al., 2009; Caporaso et al., 2010). These tools take as input large sequence data and return relatively small output files in the form of OTU relative abundance tables and OTU Fasta files. However, no formal repository or database for storing these processed datasets exists, despite the relatively small size of most of the common file types. As an essential component of addressing these questions, novel tools and computational strategies will be developed for the analysis of large biological omics datasets, to help enable an integrated analysis of crop plant microbiomes in response to drought.
Coleman-Derr D, Desgarennes D, Fonseca-Garcia C, Gross S, Clingenpeel S, Woyke T, North G, Visel A, Partida-Martinez LP, Tringe SG (2016) Plant compartment and biogeography affect microbiome composition in cultivated and native Agave species. New Phytol 209: 798–811
Singer E, Bushnell B, Coleman-Derr D, Bowman B, Levy A, Gied E, Cheng J, Copeland A, Klenk H, Hallam S, et al (2015) High-resolution phylogenetic microbial community profiling. PNAS. In Press
Wang Y, Coleman-Derr D, Chen G, Gu Y (2015) OrthoVenn: a web server for genome wide comparison and annotation of orthologous clusters across multiple species. BMC Bioinformatics.
Coleman-Derr D, Tringe SG (2014) Building the crops of tomorrow: advantages of symbiont-based approaches to improving abiotic stress tolerance. Microb Symbioses 5: 283
Martin JA, Johnson NV, Gross SM, Schnable J, Meng X, Wang M, Coleman-Derr D, Lindquist E, Wei C-L, Kaeppler S, et al (2014) A near complete snapshot of the Zea mays seedling transcriptome revealed from ultra-deep sequencing. Sci Rep. doi: 10.1038/srep04519
O’Connor RM, Fung JM, Sharp KH, Benner JS, McClung C, Cushing S, Lamkin ER, Fomenkov AI, Henrissat B, Londer YY, et al. (2014) Gill bacteria enable a novel digestive strategy in a wood-feeding mollusk. Proc Natl Acad Sci 111: E5096–E5104
Zemach A, Kim MY, Hsieh P-H, Coleman-Derr D, Eshed-Williams L, Thao K, Harmer SL, Zilberman D (2013) The Arabidopsis Nucleosome Remodeler DDM1 Allows DNA Methyltransferases to Access H1-Containing Heterochromatin. Cell 153: 193–205
Coleman-Derr D, Zilberman D (2012a) DNA Methylation, H2A.Z, and the Regulation of Constitutive Expression. Cold Spring Harb Symp Quant Biol 77: 147–154
Coleman-Derr D, Zilberman D (2012b) Deposition of Histone Variant H2A.Z within Gene Bodies Regulates Responsive Genes. PLoS Genet 8: e1002988
Akhunov ED, Akhunova AR, Anderson OD, Anderson JA, Blake N, Clegg MT, Coleman-Derr D, Conley EJ, Crossman CC, Deal KR, et al (2010) Nucleotide diversity maps reveal variation in diversity among wheat genomes and chromosomes. BMC Genomics 11: 702
Gu YQ, Wanjugi H, Coleman-Derr D, Kong X, Anderson OD (2010) Conserved globulin gene across eight grass genomes identify fundamental units of the loci encoding seed storage proteins. Funct Integr Genomics 10: 111–122
Wanjugi H, Coleman-Derr D, Huo N, Kianian SF, Luo M-C, Wu J, Anderson O, Gu YQ (2009) Rapid development of PCR-based genome-specific repetitive DNA junction markers in wheat. Genome 52: 576–587
Zilberman D, Coleman-Derr D, Ballinger T, Henikoff S (2008) Histone H2A.Z and DNA methylation are mutually antagonistic chromatin marks. Nature 456: 125–129
Gao S, Gu YQ, Wu J, Coleman-Derr D, Huo N, Crossman C, Jia J, Zuo Q, Ren Z, Anderson OD, et al (2007) Rapid evolution and complex structural organization in genomic regions harboring multiple prolamin genes in the polyploid wheat genome. Plant Mol Biol 65: 189–203
Zhang S, Gu YQ, Singh J, Coleman-Derr D, Brar DS, Jiang N, Lemaux PG (2007) New insights into Oryza genome evolution: high gene colinearity and differential retrotransposon amplification. Plant Mol Biol 64: 589–600
Gu YQ, Salse J, Coleman-Derr D, Dupin A, Crossman C, Lazo GR, Huo N, Belcram H, Ravel C, Charmet G, et al (2006) Types and Rates of Sequence Evolution at the High-Molecular-Weight Glutenin Locus in Hexaploid Wheat and Its Ancestral Genomes. Genetics 174: 1493–1504
Huo N, Gu YQ, Lazo GR, Vogel JP, Coleman-Derr D, Luo M-C, Thilmony R, Garvin DF, Anderson OD (2006) Construction and characterization of two BAC libraries from Brachypodium distachyon, a new model for grass genomics. Genome 49: 1099–1108
Vogel JP, Gu YQ, Twigg P, Lazo GR, Laudencia-Chingcuanco D, Hayden DM, Donze TJ, Vivian LA, Stamova B, Coleman-Derr D (2006) EST sequencing and phylogenetic analysis of the model grass Brachypodium distachyon. Theor Appl Genet 113: 186–195
Devin Coleman-Derr
Albany, CA